Cancer Mutation
Dongyue Xie
2017-07-17
Last updated: 2017-09-14
Code version: be155e6
Detection of cancer mutaiton signatures is important in the study of the causes and mechanism of cancers. Usually we consider 6 possible mutation patterns(C to A/G/T, T to A/C/G). Shiraishi et al.(2015) proposed a parsimonious approach to model the mutation signatures, by assuming independence across mutation patterns. Here, we use logo plots to visualize the mutaiton signatures from two data sets: Shiraishi et al.(2015) and Alexandrov et al.(2013).
Shiraishi et al.(2015)
Depletion cases: 26, C at +1; 15,16,25 G at +1
Depletion of subsitituion: 22,23,24, with G,C depleted at flanking sites.
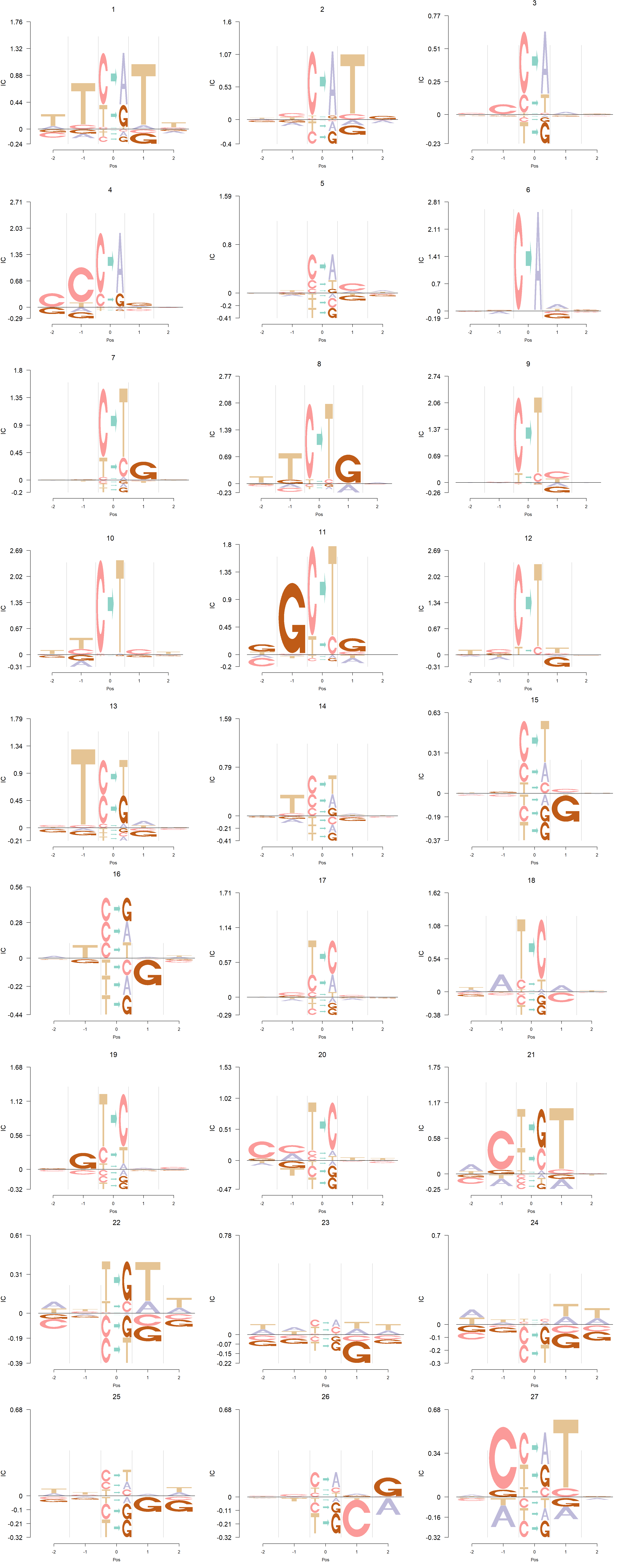
The logo height below is the “Log-odds Height”.
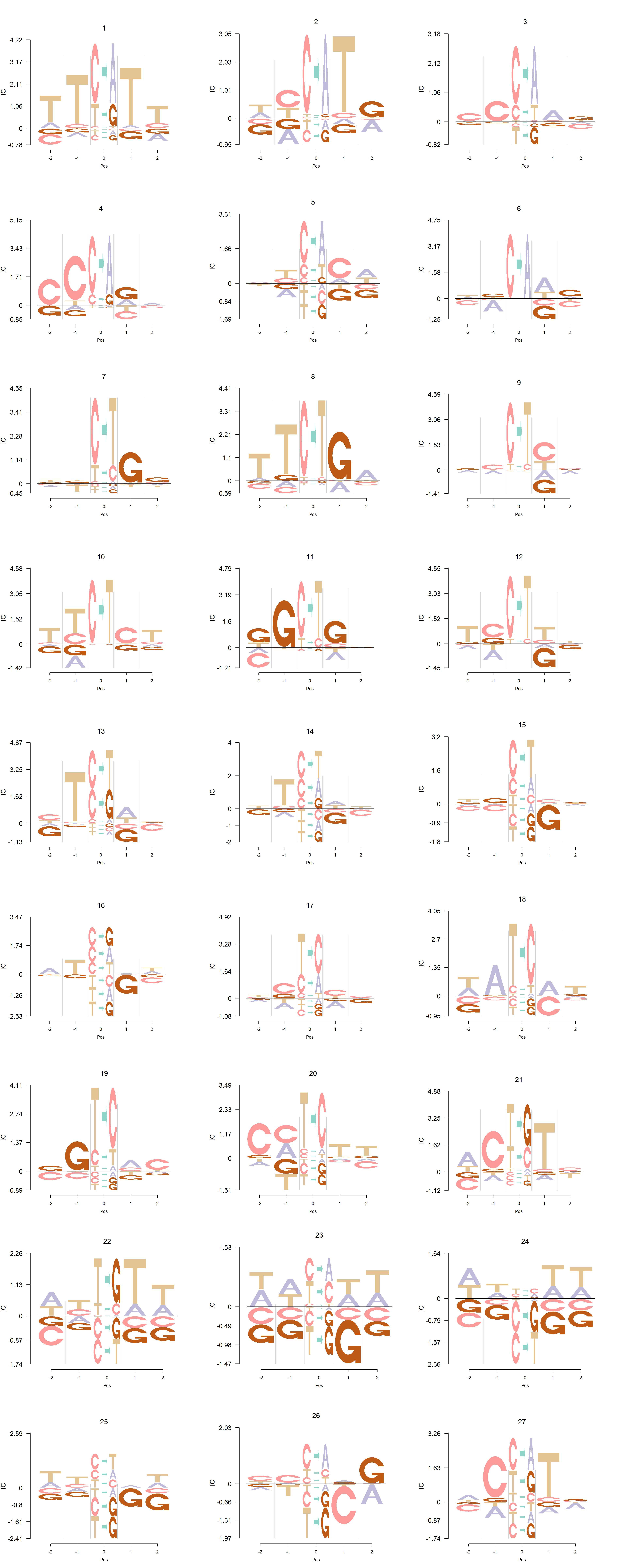
Alexandrov et al.(2013).
In Shiraishi et al.(2015), the Supp Fig 3 lists several signatures extracted in each cancer type in the Alexandrov et al. (2013) data. Here, we compare the signature from Logolas and Shiraishi.
Cancer mutation data are from here.
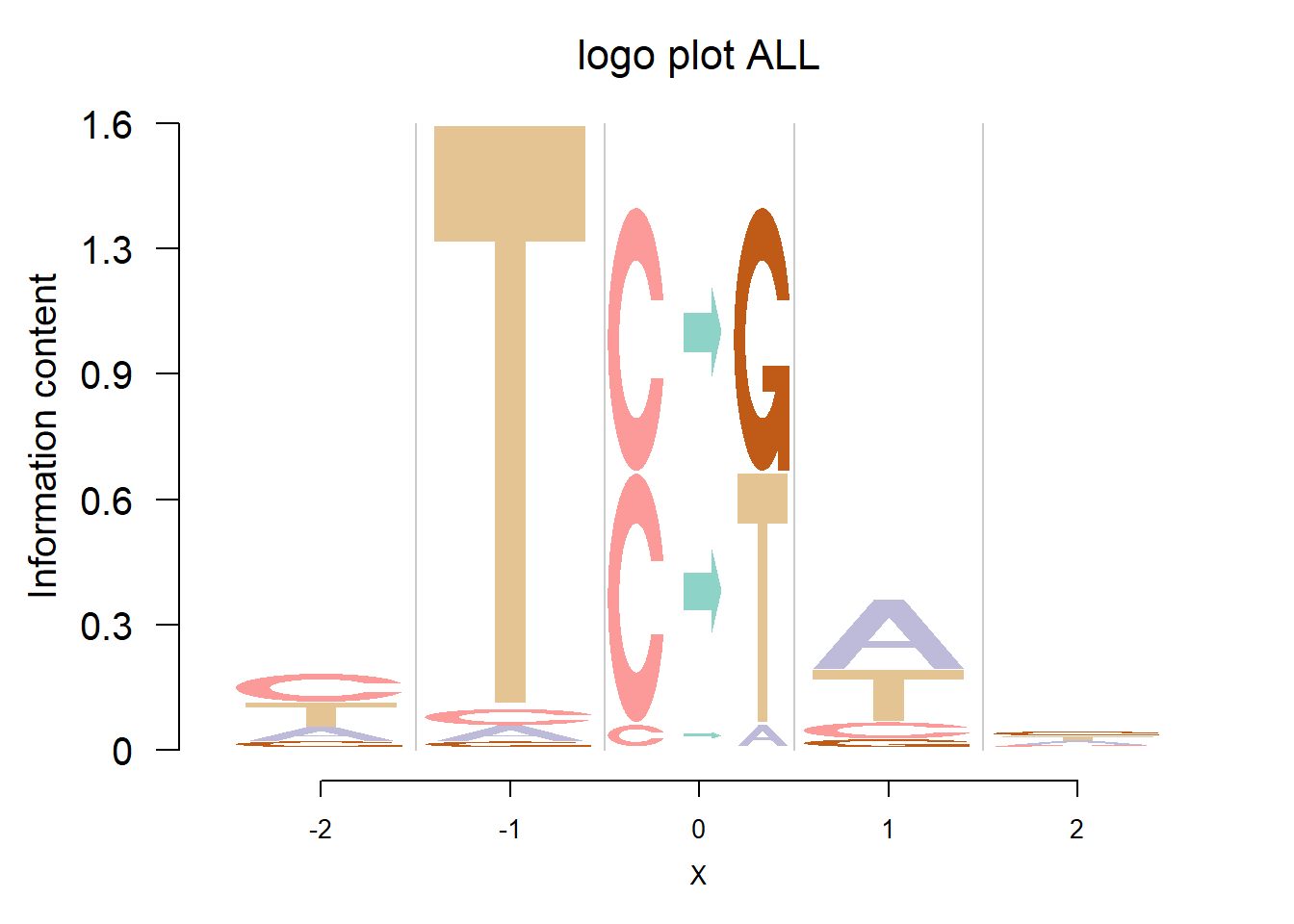
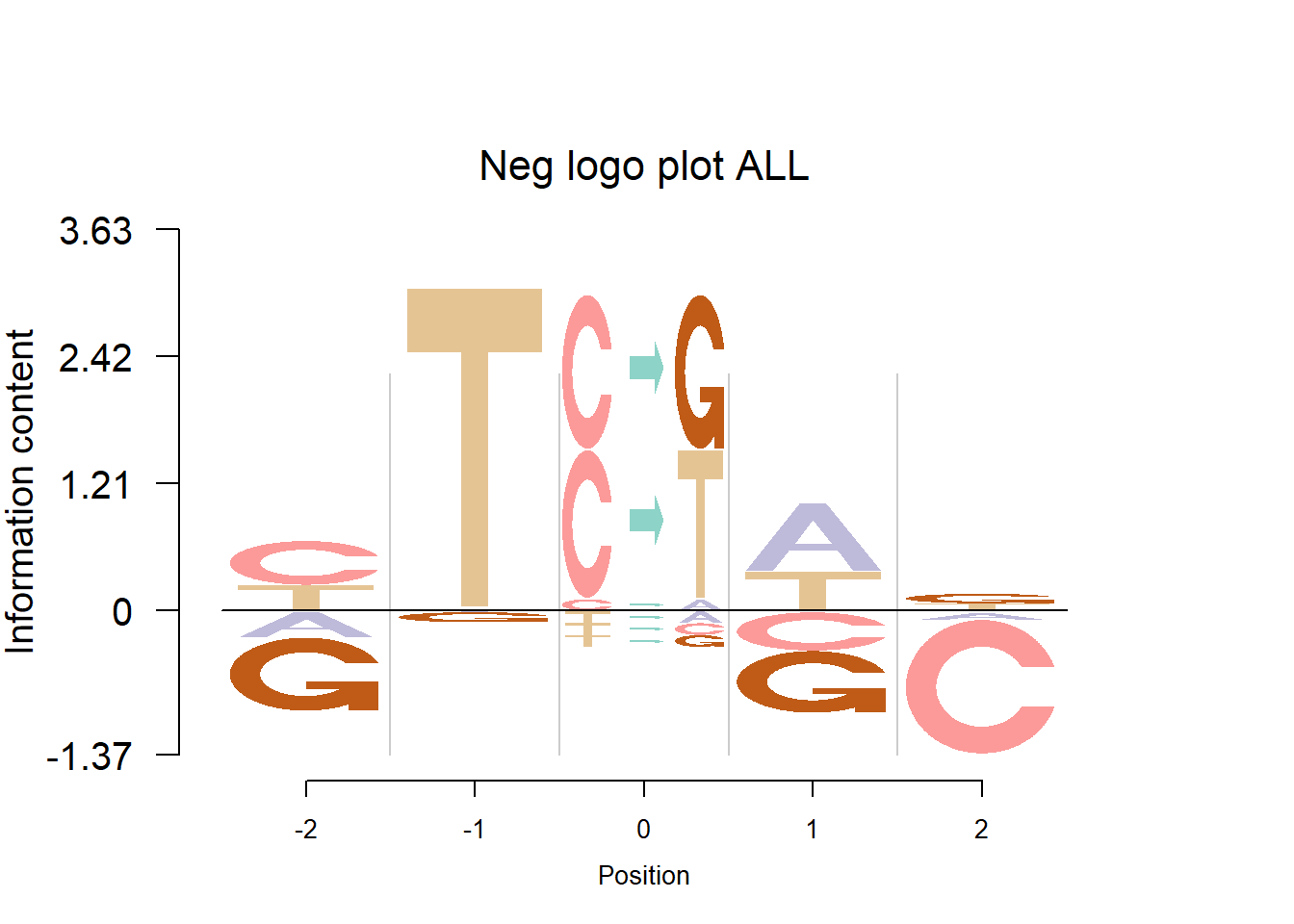
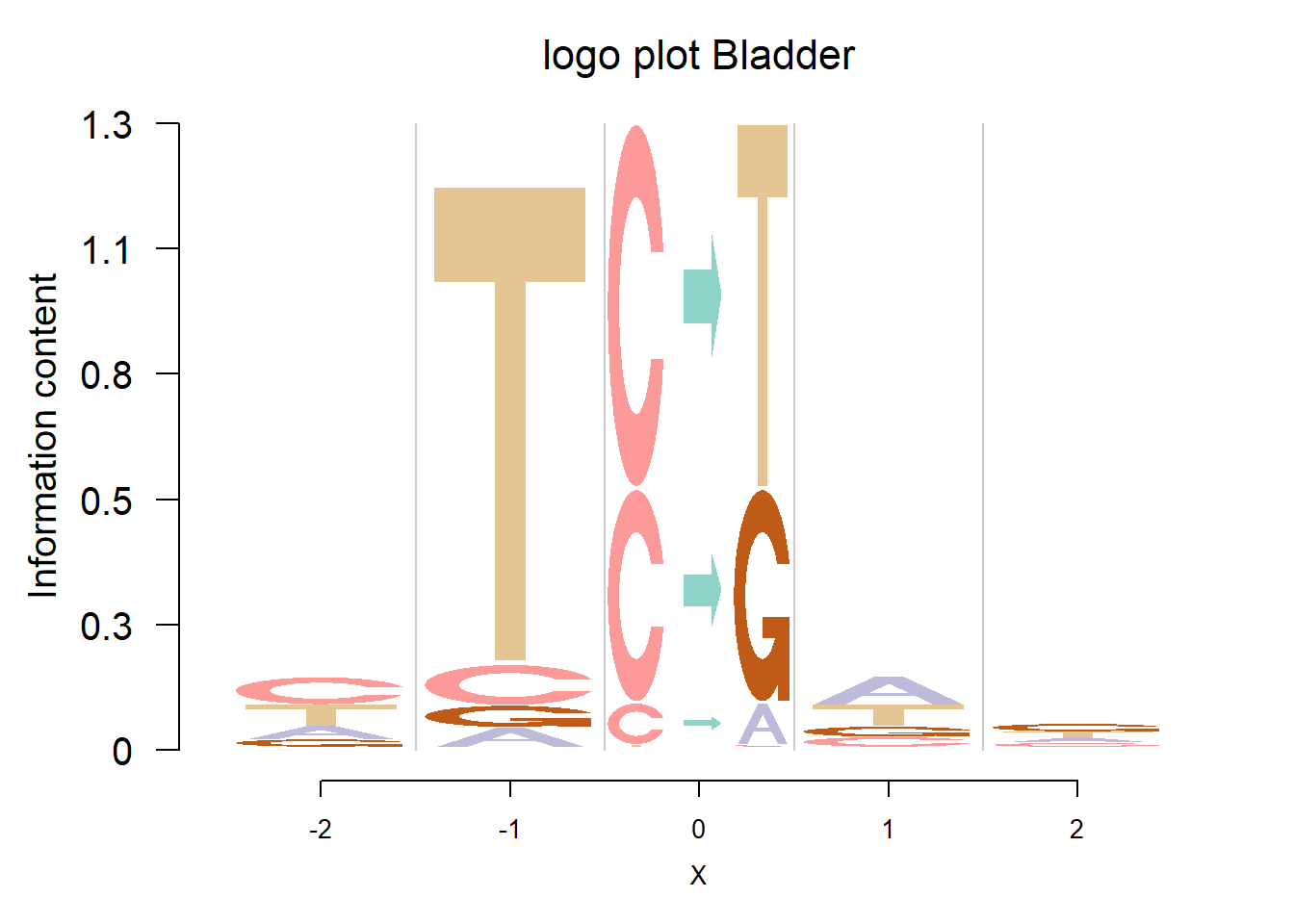
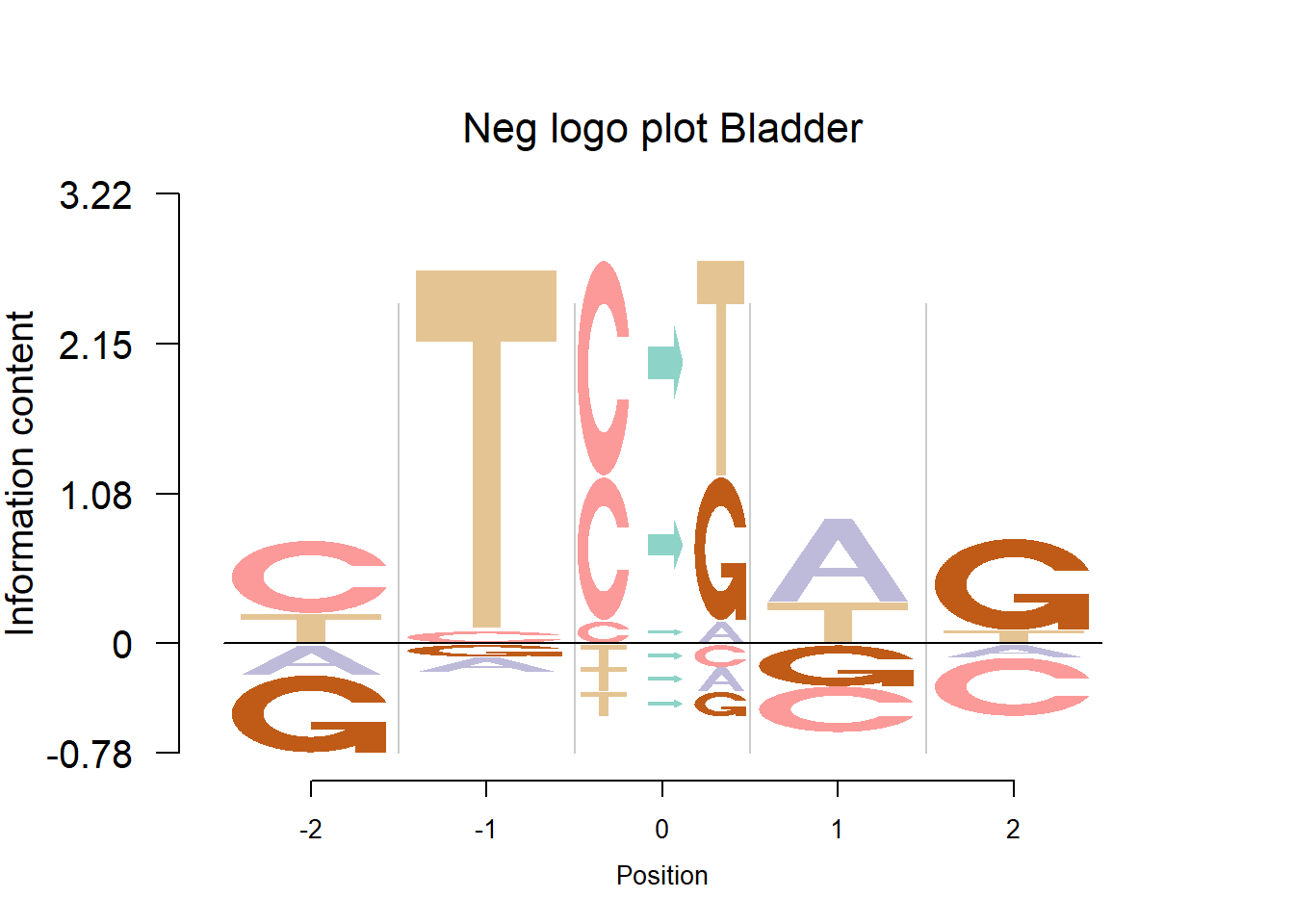
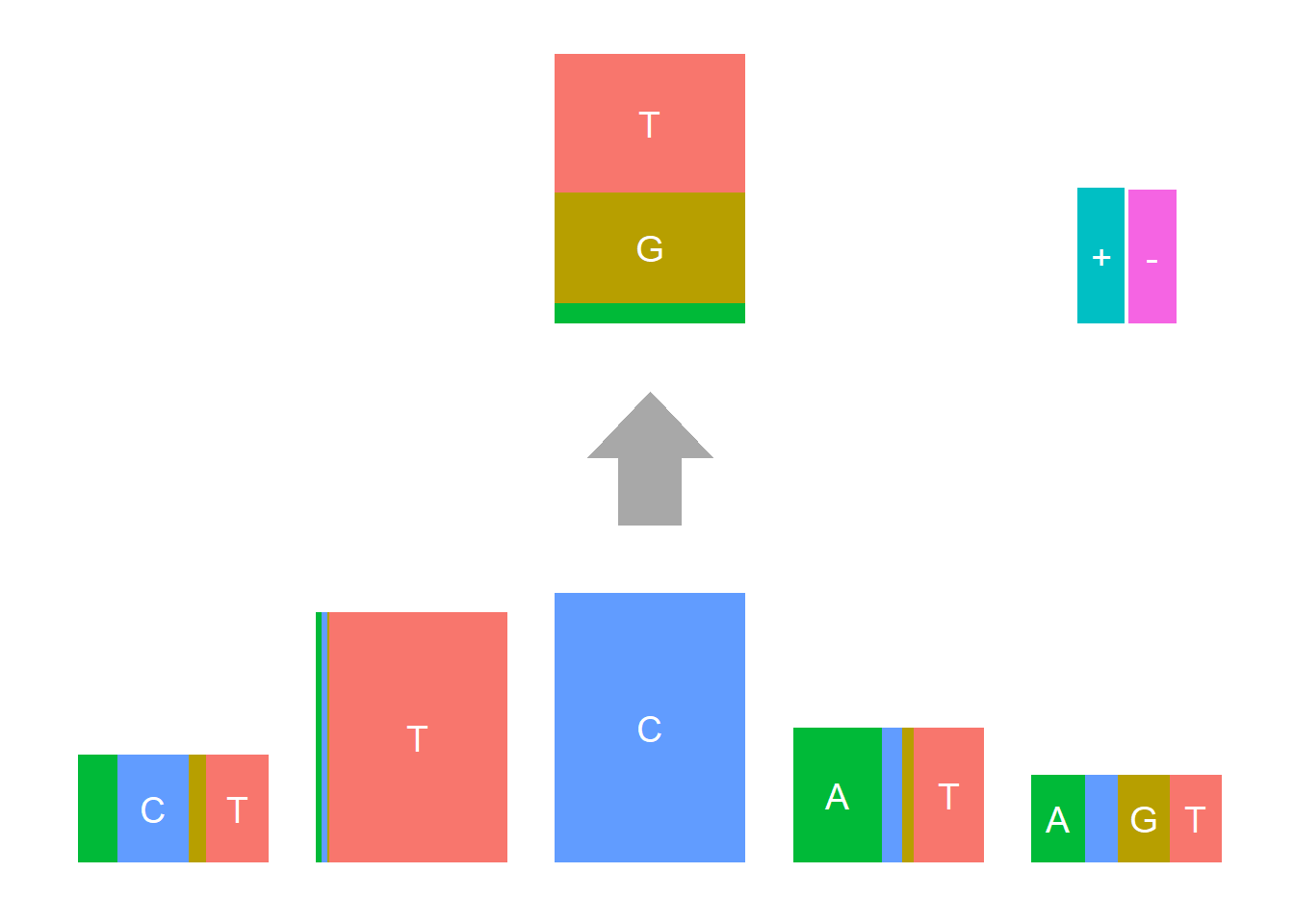
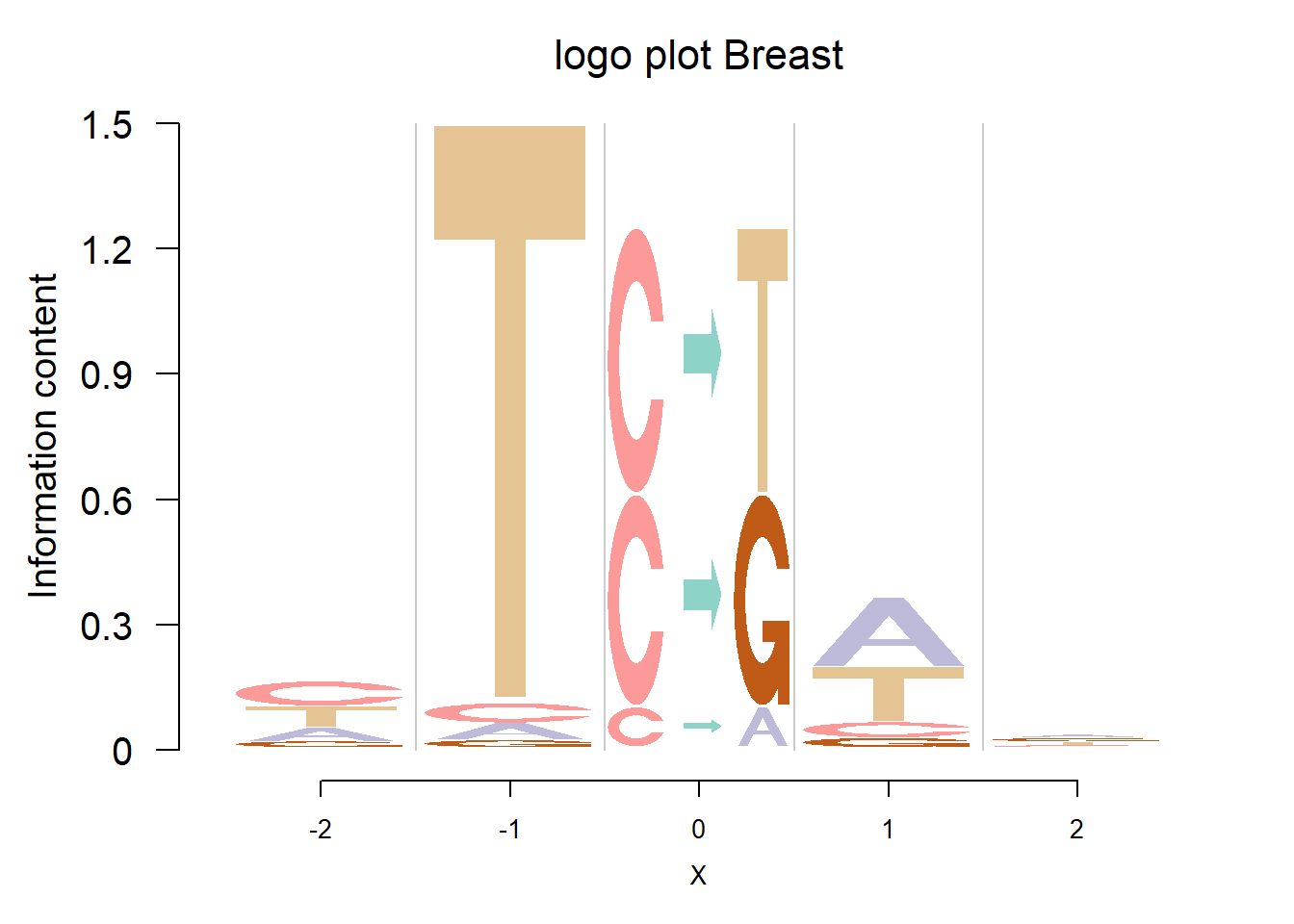
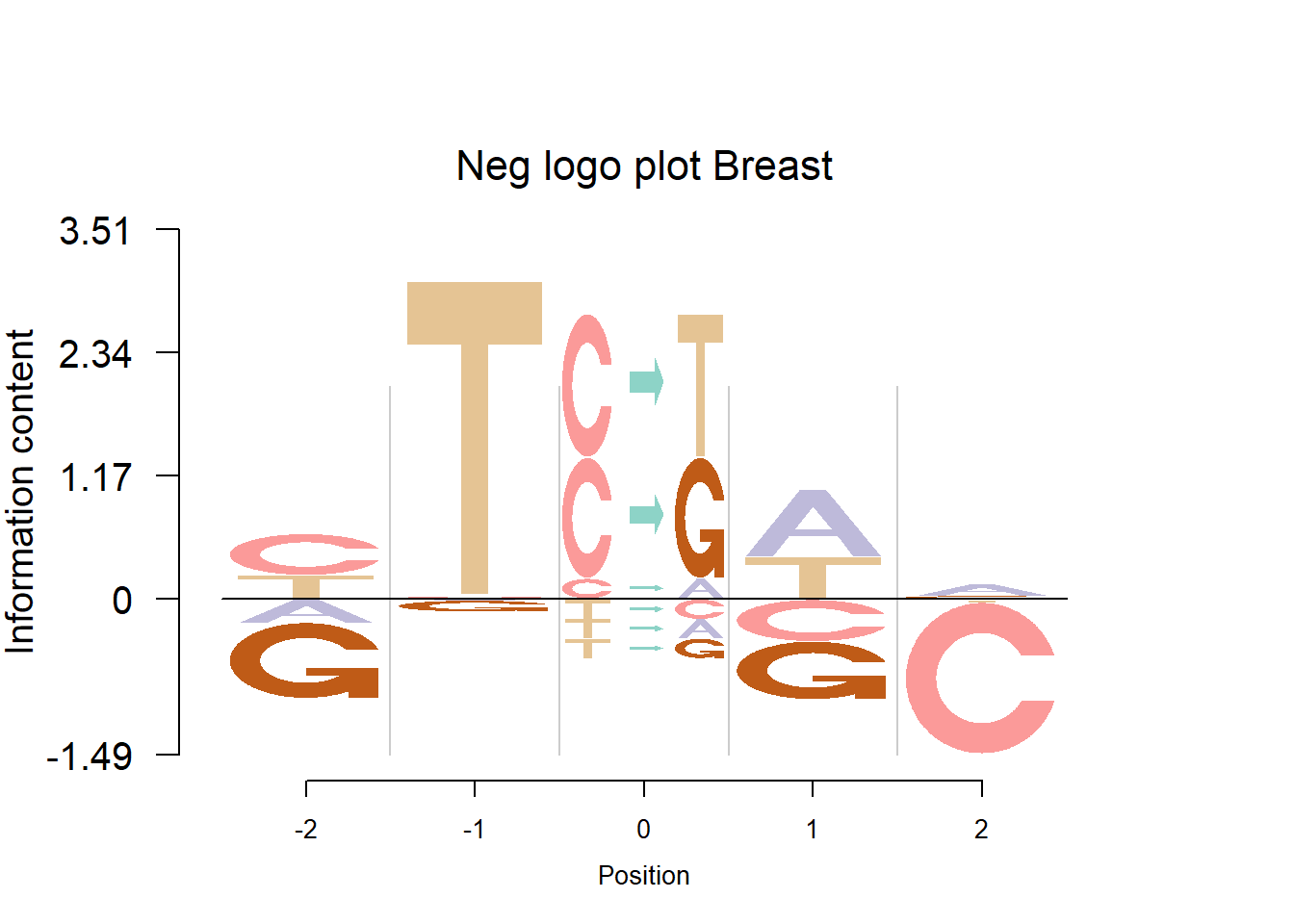
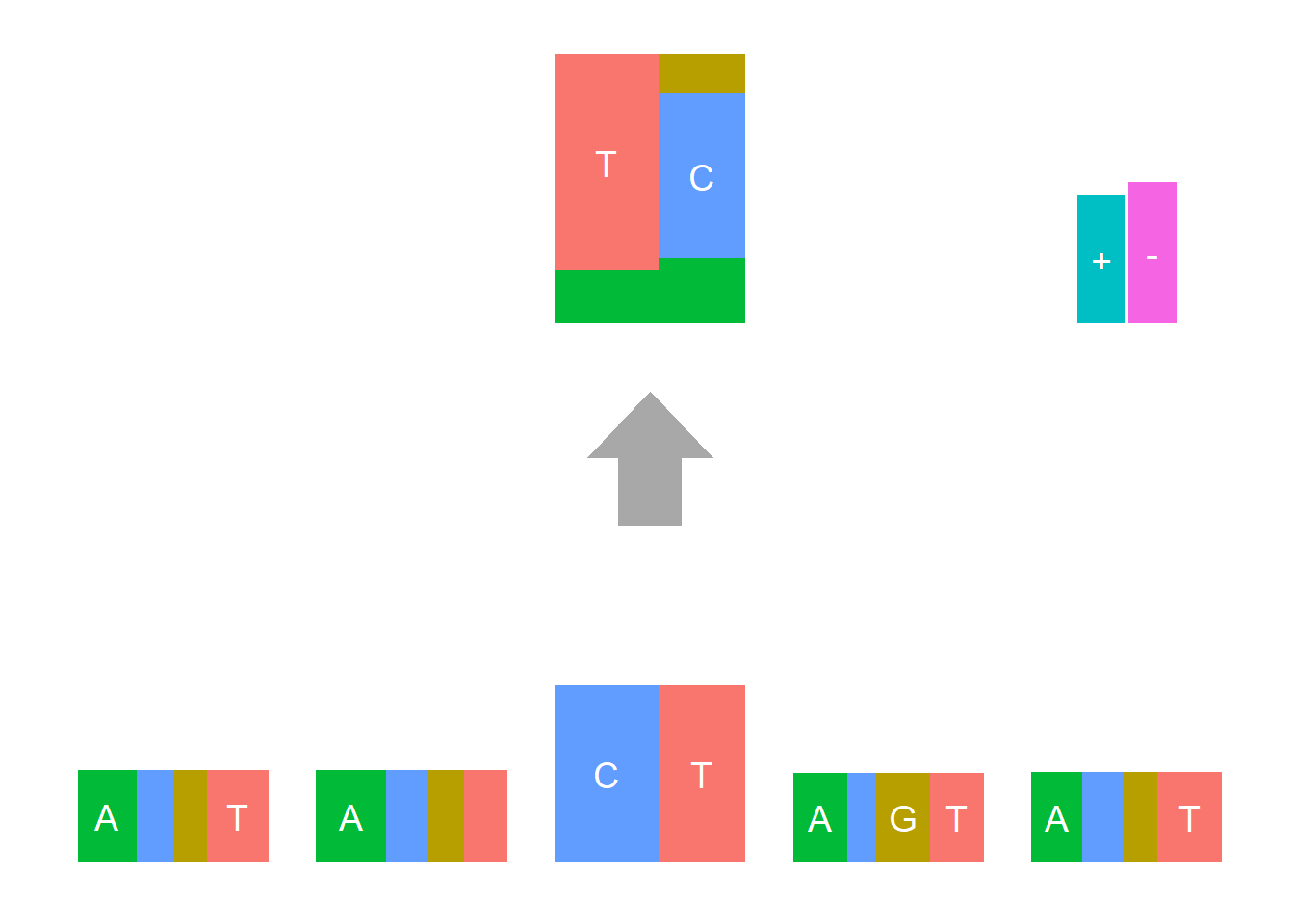
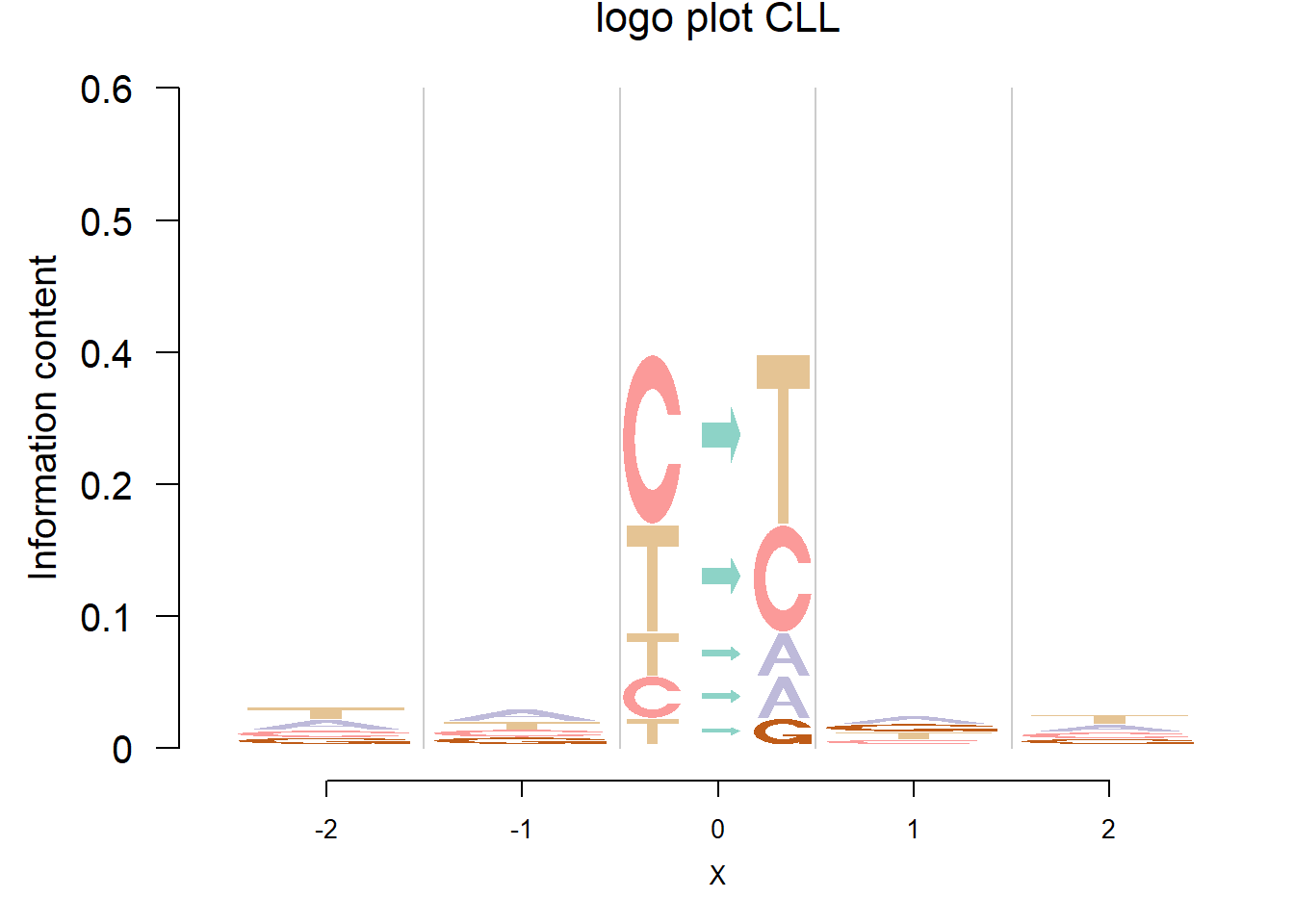
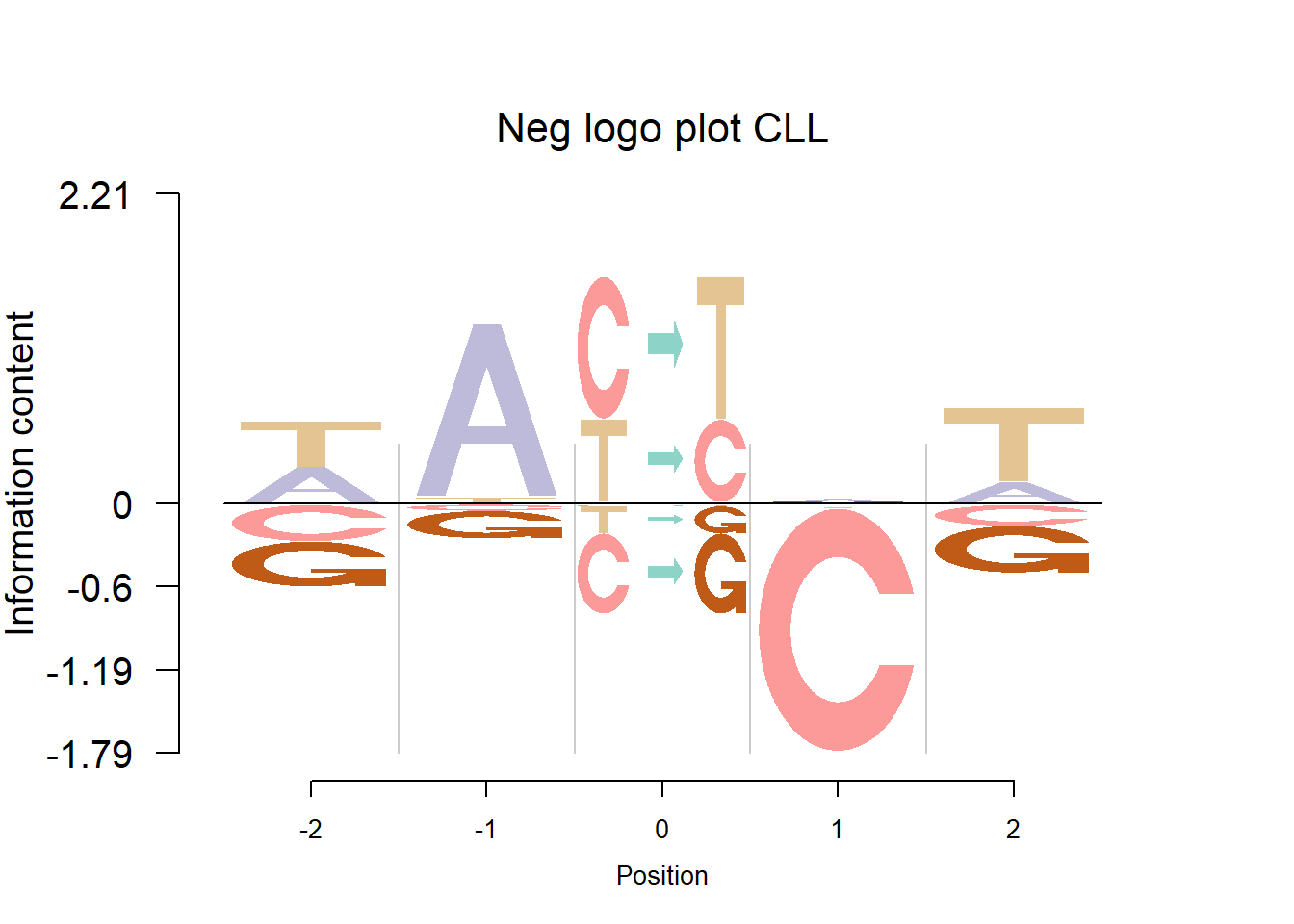
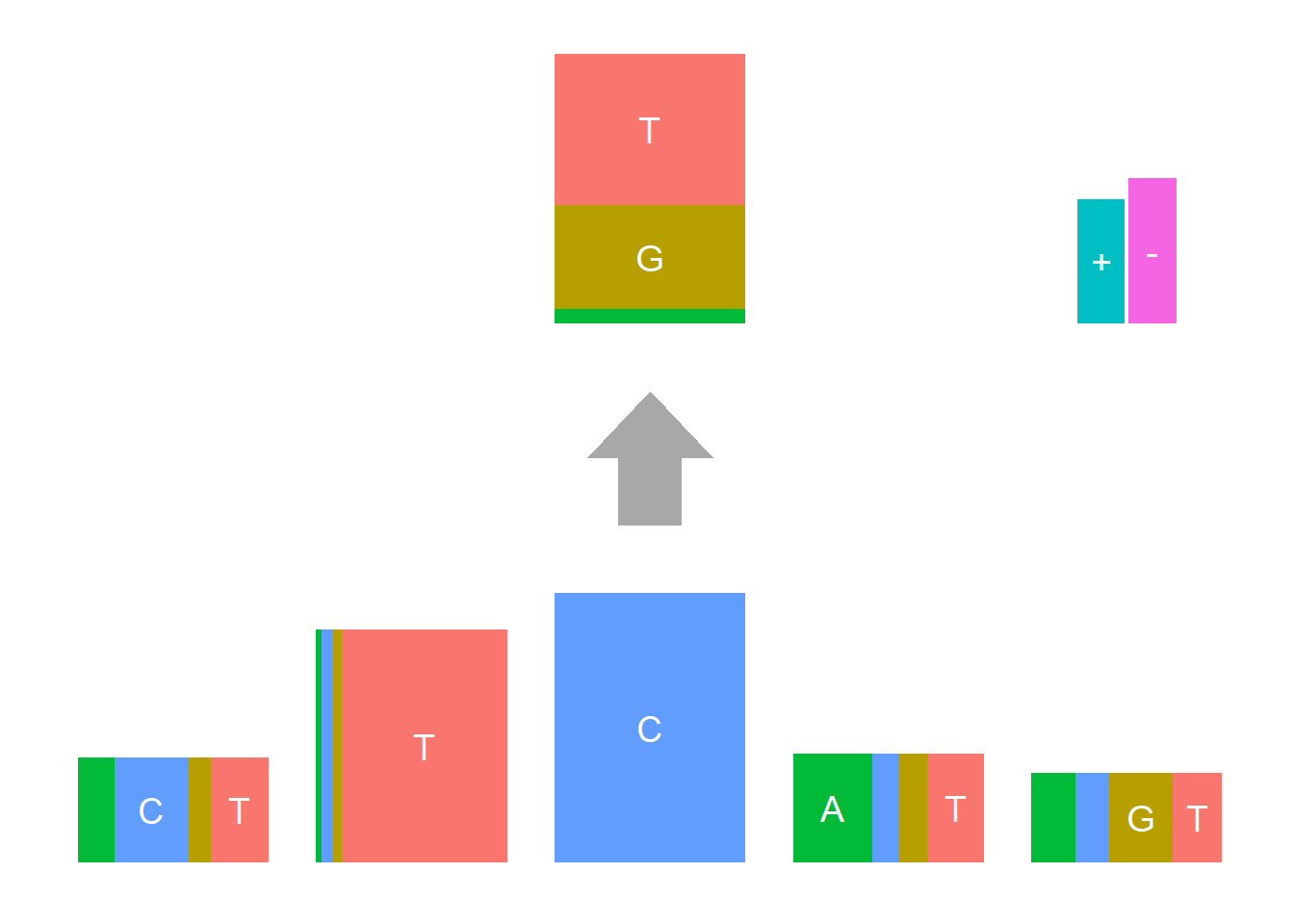
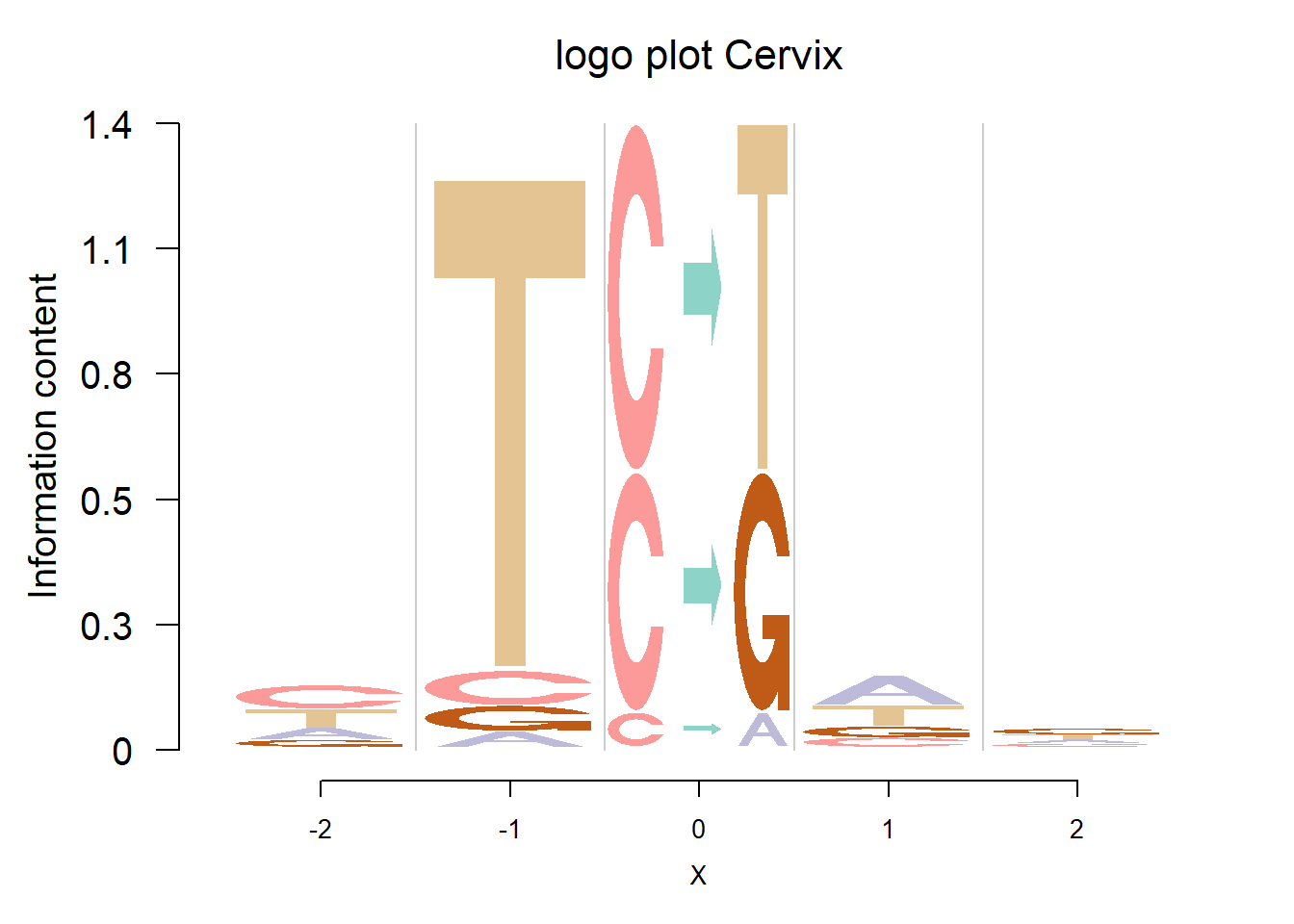
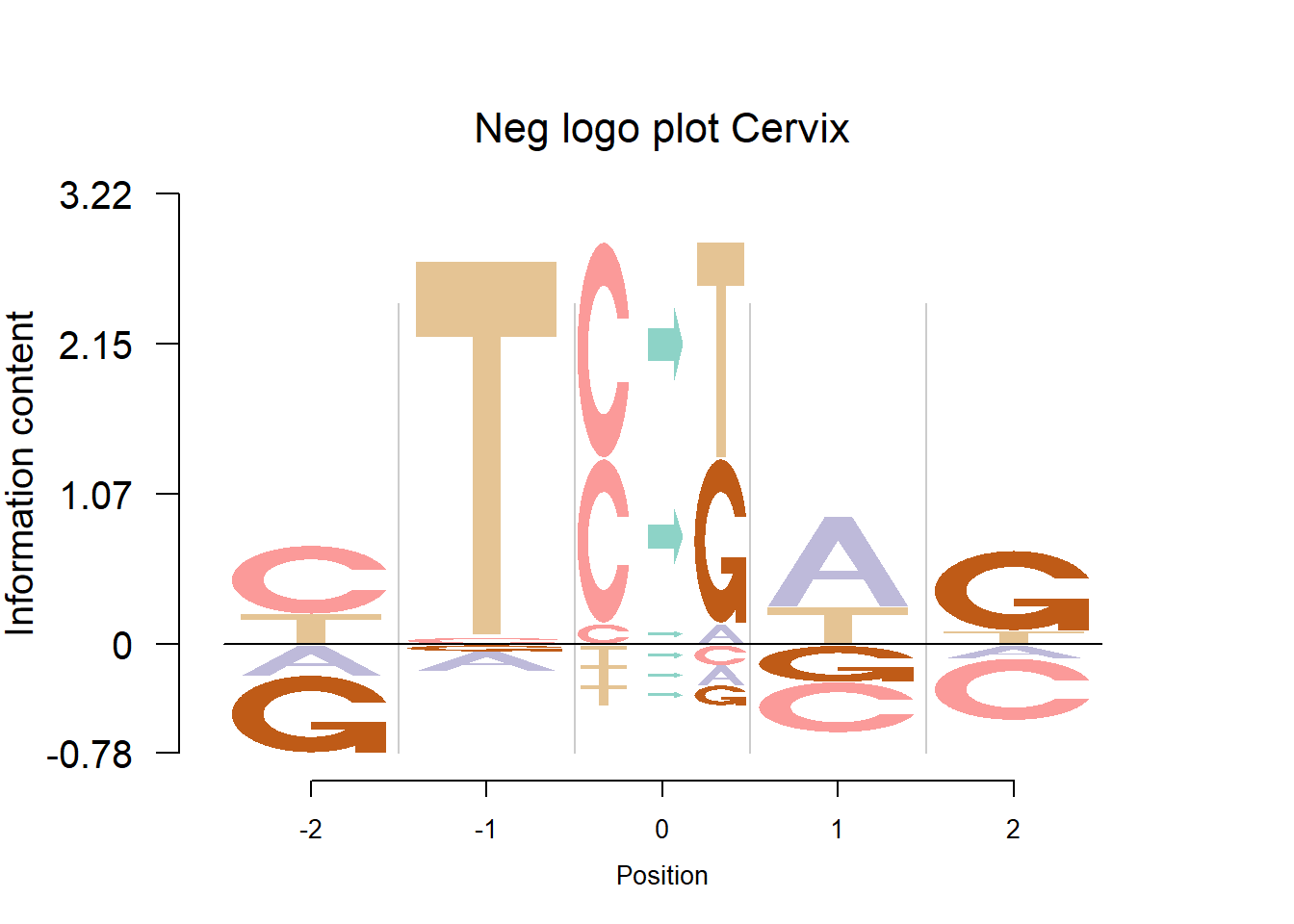
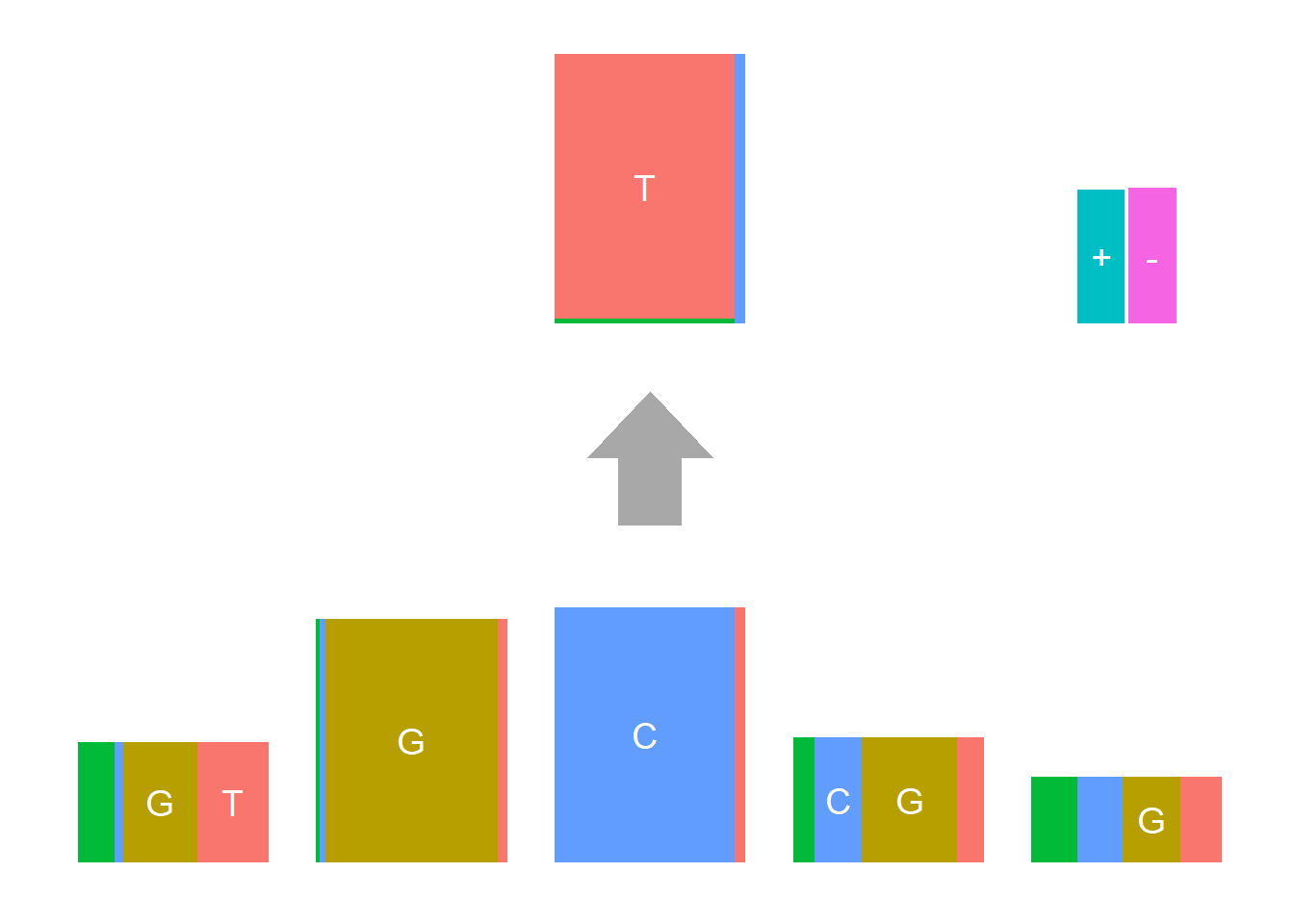
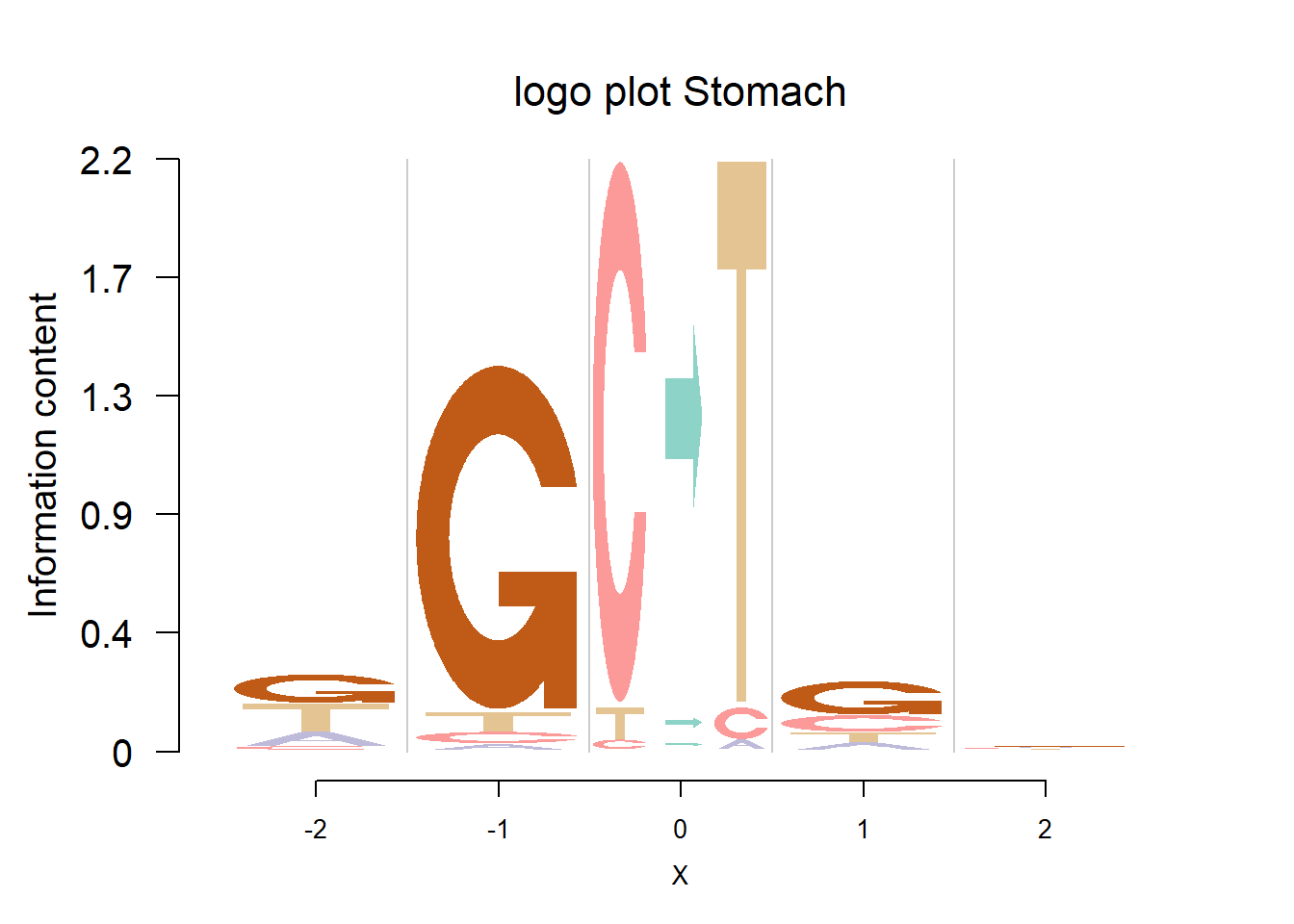
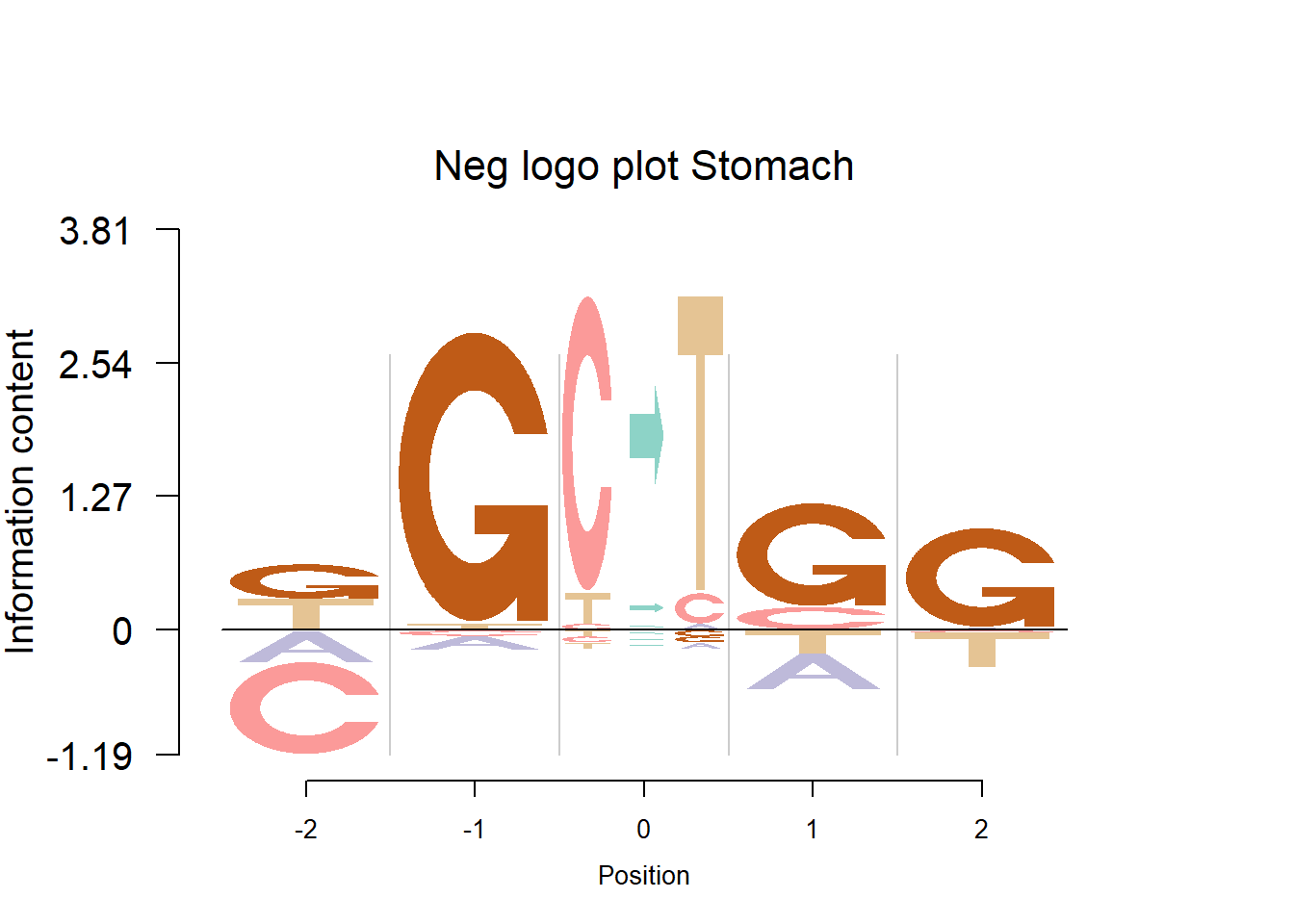
The Supp Fig 3 in the original paper Shiraishi et al.(2015) is attached below.

Reference
Shiraishi, Y., Tremmel, G., Miyano, S., & Stephens, M. (2015). A simple model-based approach to inferring and visualizing cancer mutation signatures. PLoS genetics, 11(12), e1005657.
Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 2013 Jan;3(1):246-259. pmid:23318258
Session information
sessionInfo()R version 3.4.0 (2017-04-21)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 15063)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.1252
[2] LC_CTYPE=English_United States.1252
[3] LC_MONETARY=English_United States.1252
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.1252
attached base packages:
[1] grid stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] pmsignature_0.3.0 tidyr_0.6.3 dplyr_0.7.2 ggplot2_2.2.1
[5] Logolas_1.1.2
loaded via a namespace (and not attached):
[1] Rcpp_0.12.12 bindr_0.1 knitr_1.15.1
[4] magrittr_1.5 munsell_0.4.3 SQUAREM_2016.8-2
[7] colorspace_1.3-2 R6_2.2.0 rlang_0.1.1
[10] stringr_1.2.0 plyr_1.8.4 tools_3.4.0
[13] parallel_3.4.0 gtable_0.2.0 git2r_0.18.0
[16] htmltools_0.3.5 assertthat_0.2.0 lazyeval_0.2.0
[19] yaml_2.1.14 rprojroot_1.2 digest_0.6.12
[22] tibble_1.3.3 bindrcpp_0.2 RColorBrewer_1.1-2
[25] glue_1.1.1 evaluate_0.10 rmarkdown_1.6
[28] LaplacesDemon_16.0.1 labeling_0.3 stringi_1.1.5
[31] compiler_3.4.0 scales_0.4.1 backports_1.0.5
[34] pkgconfig_2.0.1 This R Markdown site was created with workflowr